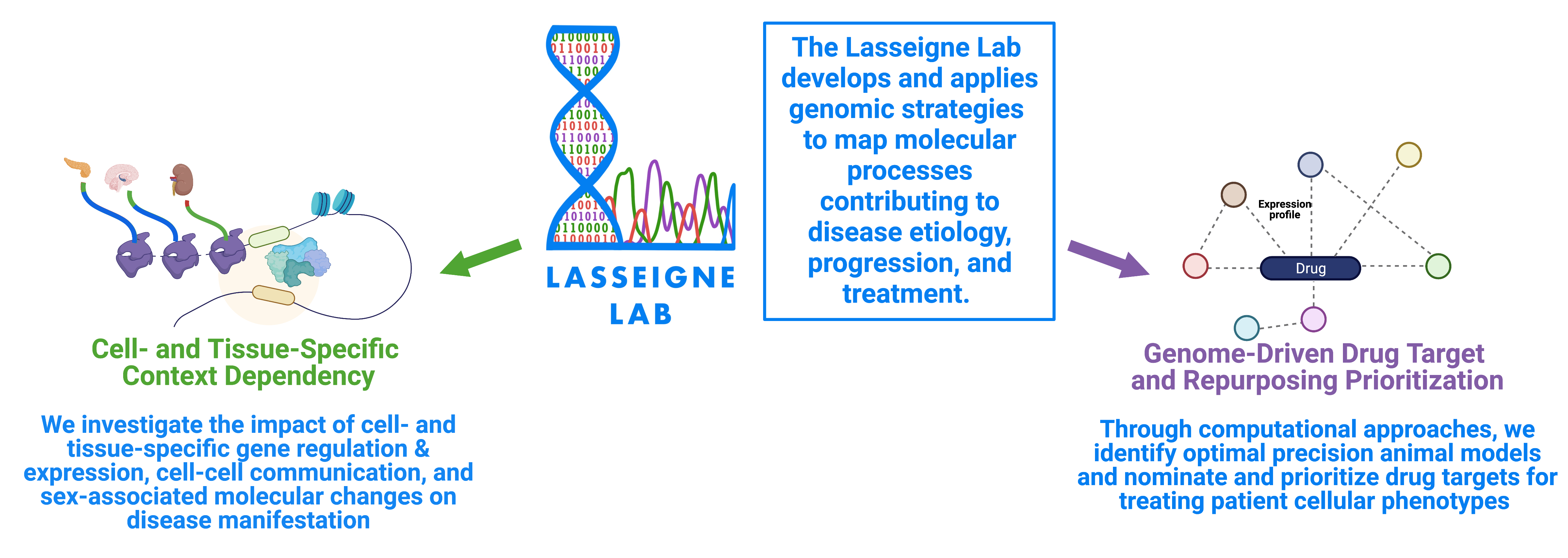
The Lasseigne Lab develops and applies genomic strategies to map molecular processes contributing to the etiology, progression, and treatment of diseases originating in the brain and/or the kidney. We investigate the impact of cell- and tissue-specific gene and transcript regulation and expression, cell-cell communication, and sex-associated molecular changes on disease manifestation. Through computational machine learning and data-driven approaches, we identify optimal precision preclinical models and nominate and prioritize drug targets and repositioning candidates for treating patient cellular phenotypes.
Some of the questions our lab asks are: If Mendelian disease associated variants are present in every cell of the body, what cellular environments and mechanisms determine why some cells show a molecular or clinical phenotype when others do not? Why are some people or some tissues more susceptible to adverse events after taking medications than others? How does differential transcript expression impact disease progression? Can we identify gene regulatory states more susceptible to treatments and induce those states in cells? Which ligand-receptor-gene target relationship changes are important for disease progression and/or treatment?
Kidney Research
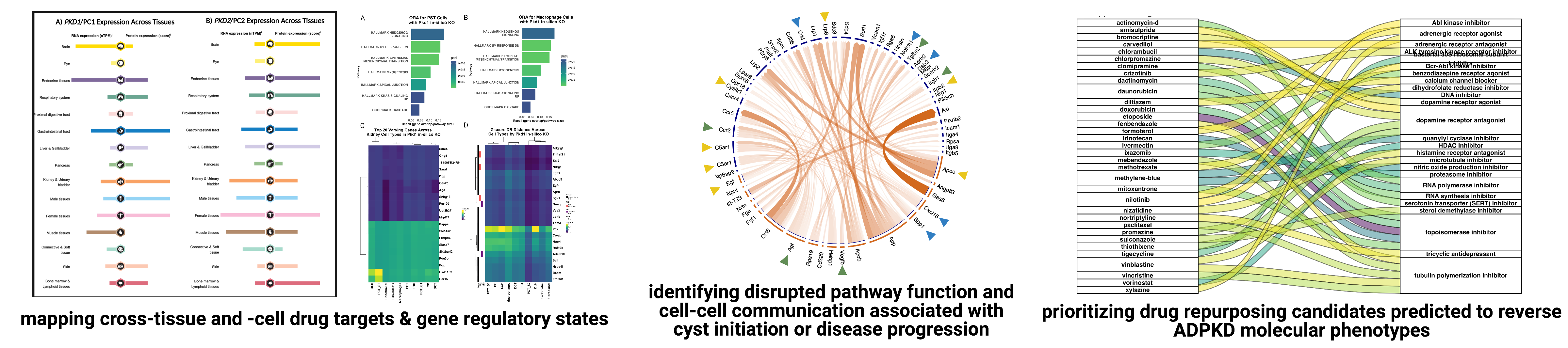
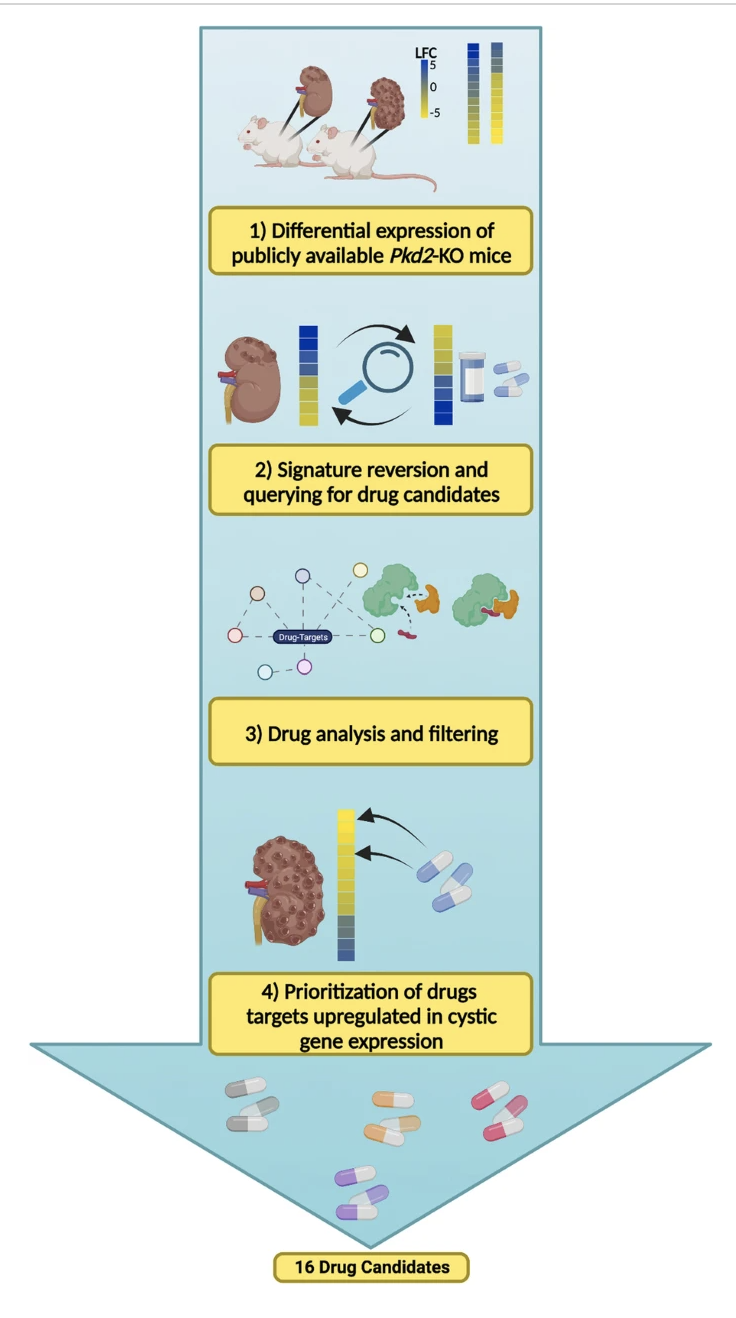
The Lasseigne Lab is currently engaged in multiple collaborative projects aimed at understanding the role of cell-cell communication, acute kidney injury, and cell state in polycystic kidney disease progression and how macrophage subpopulations respond to acute kidney injury with the goal of identifying disease mechanisms and potential drug targets for both through genomics approaches.
Recent publications/preprints:
Brain Research
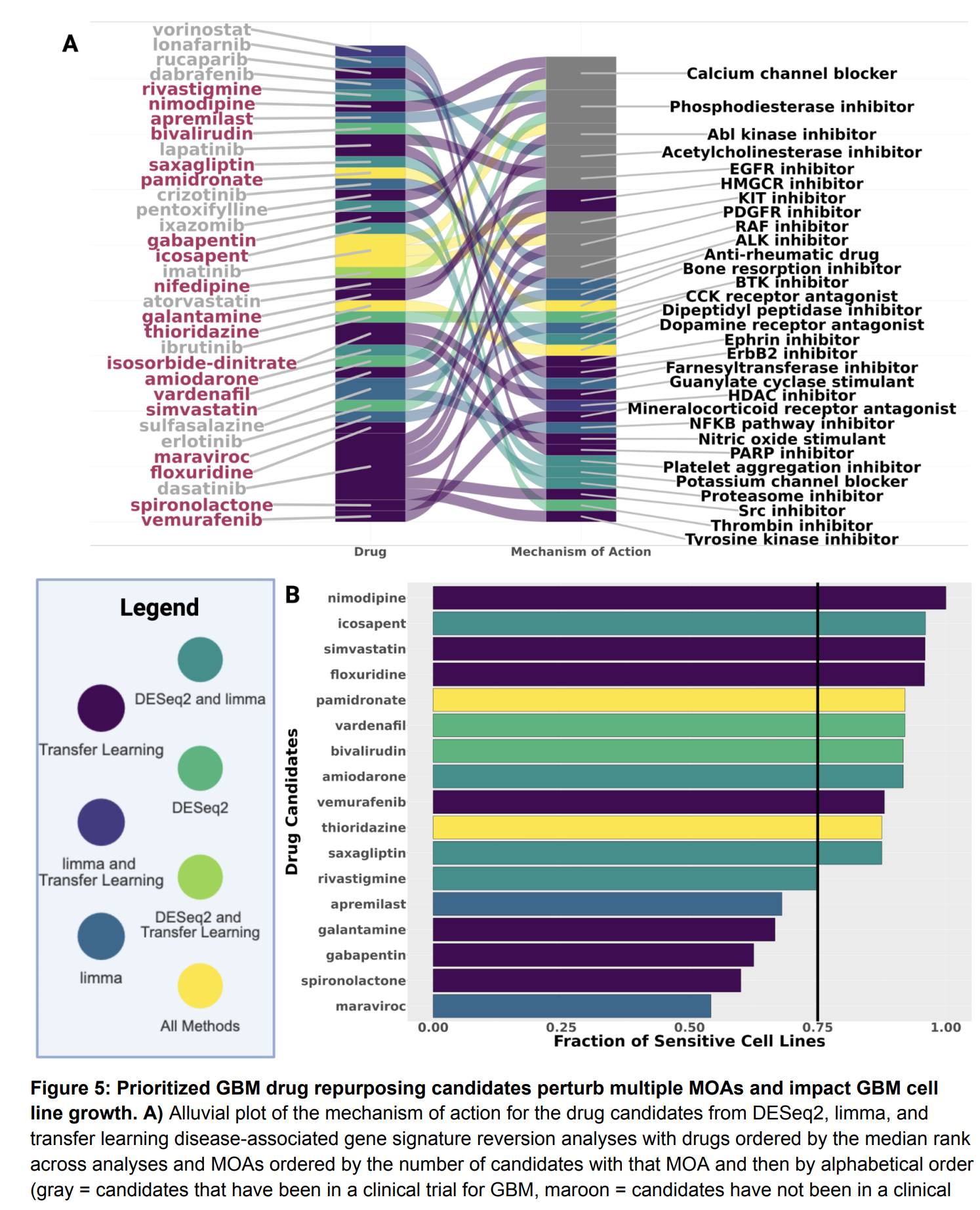
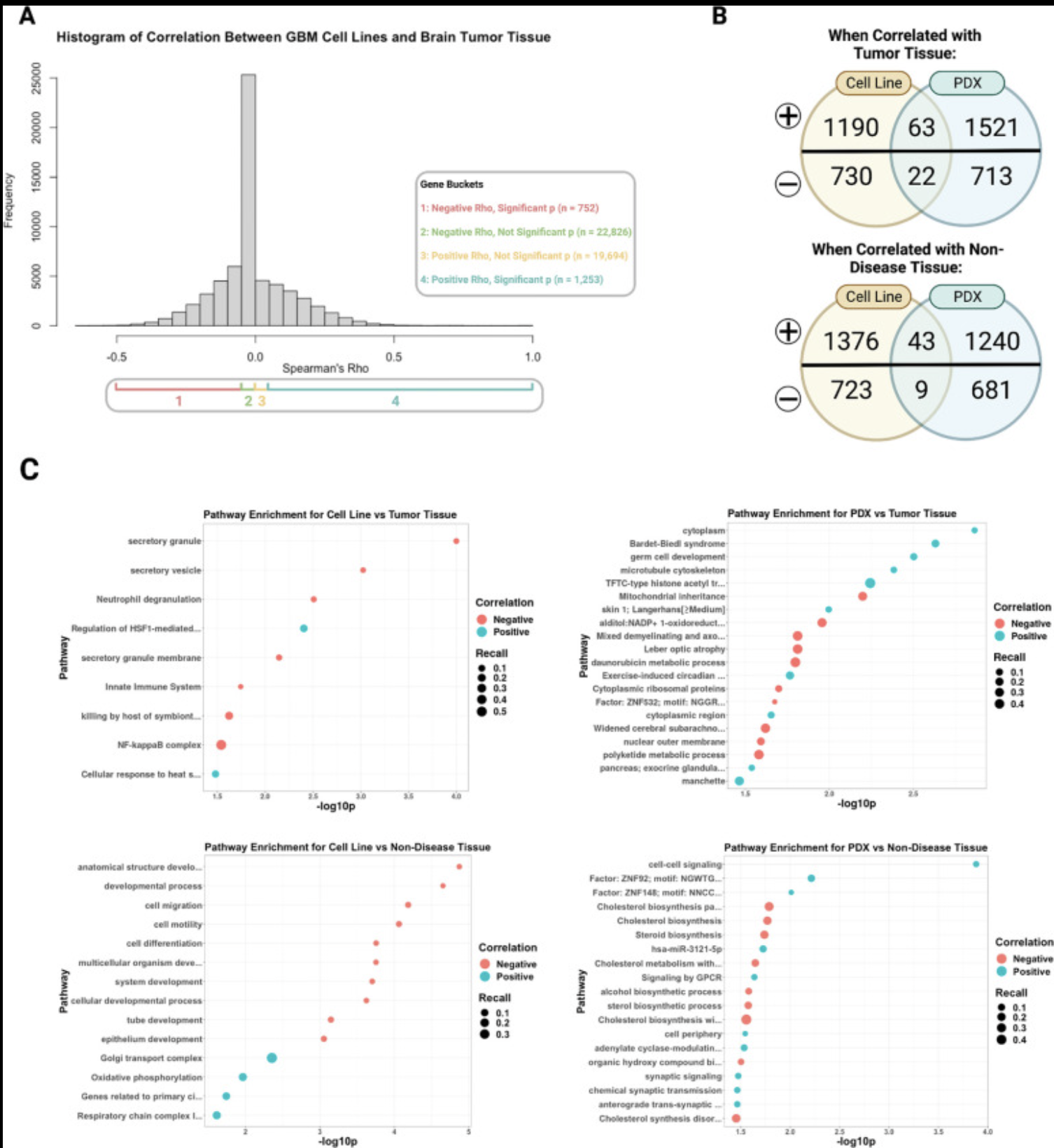
Our lab has several collaborative projects leveraging single-cell and long-read genomics approaches to learn how different cell types contribute to Alzheimer’s, glioblastoma, and rare diseases impacting the brain through changes in cell-cell communication, transcription factor activity, and gene regulatory states.
Recent publications/preprints:
Drug Target and Repurposing Prioritization
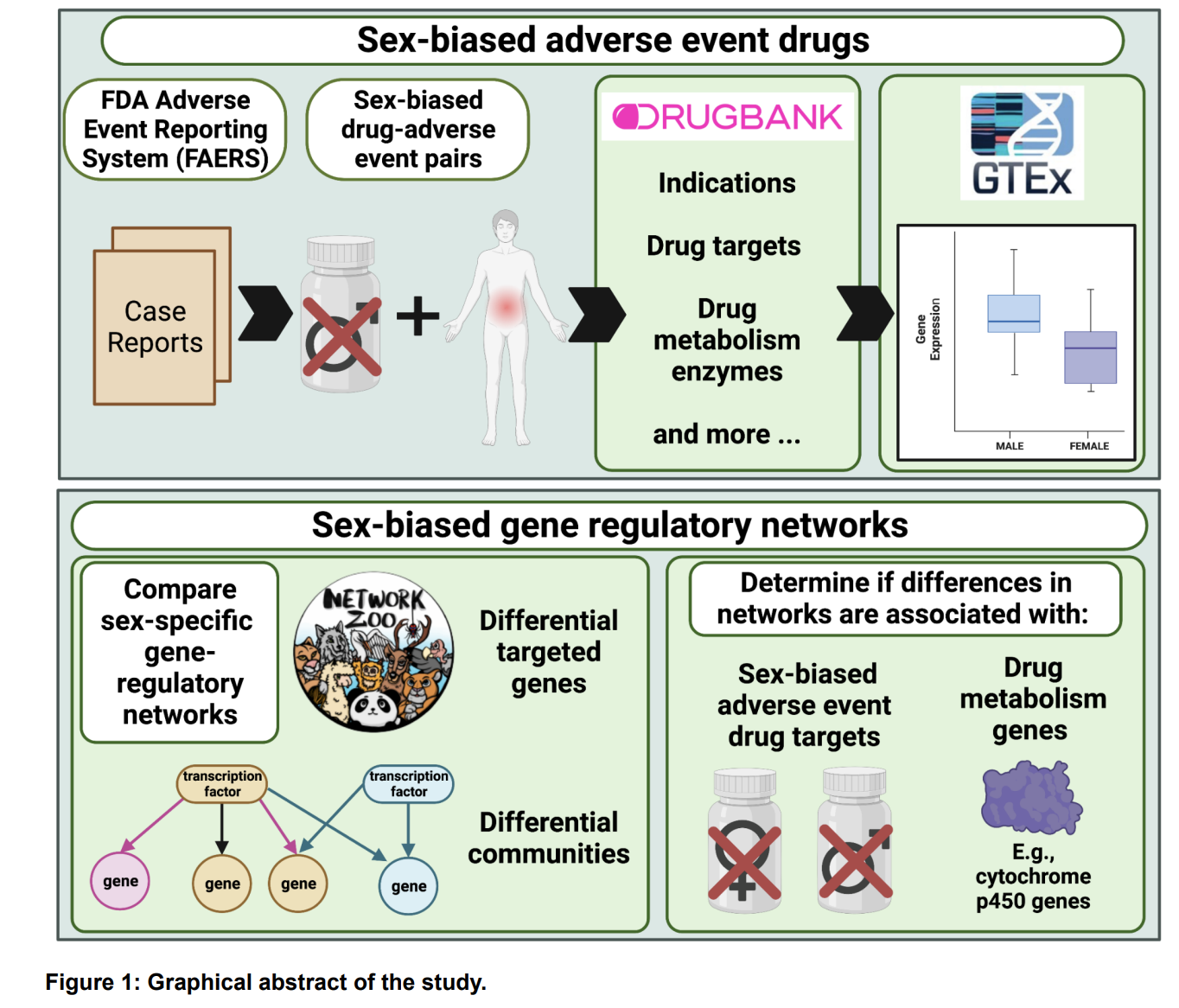
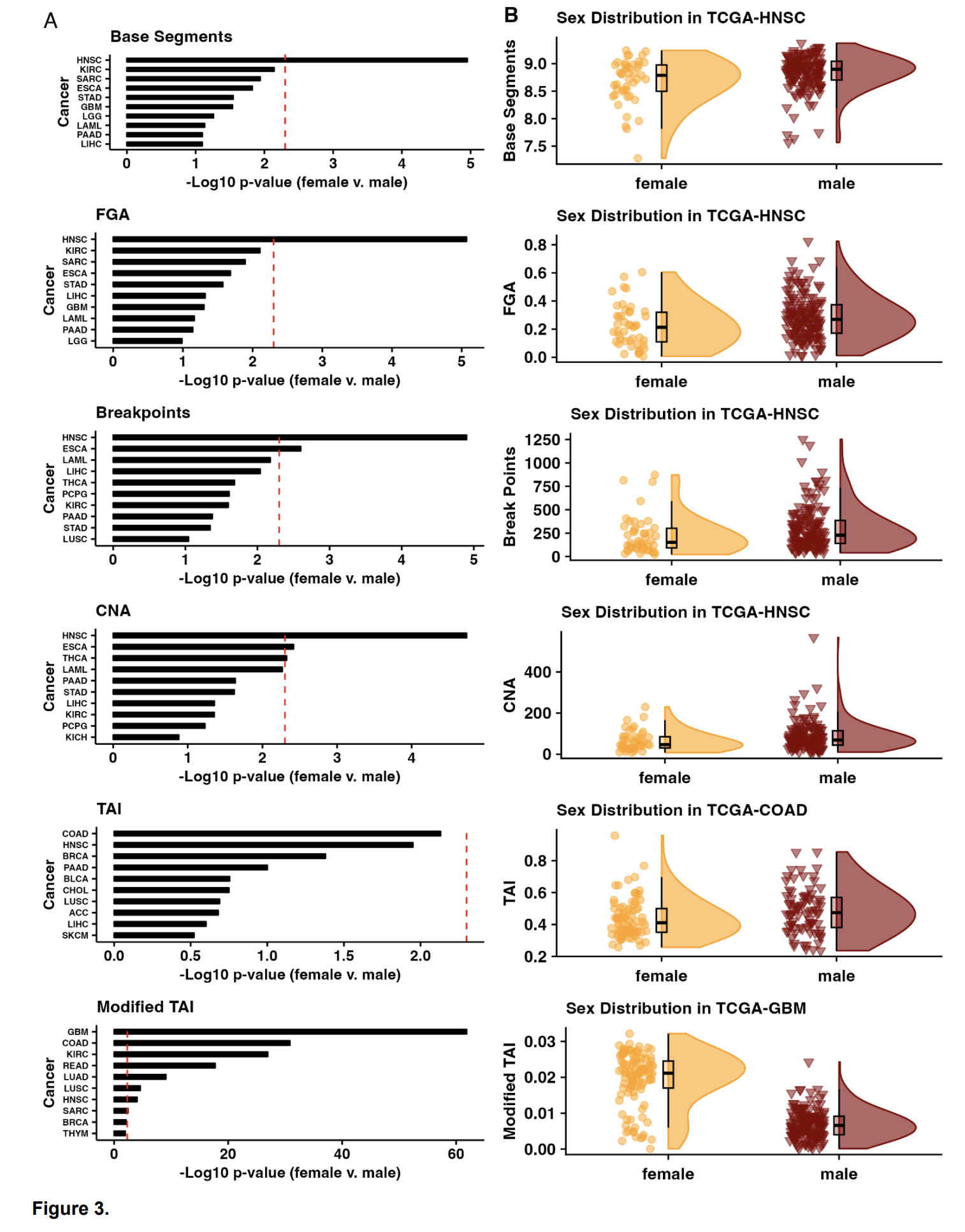
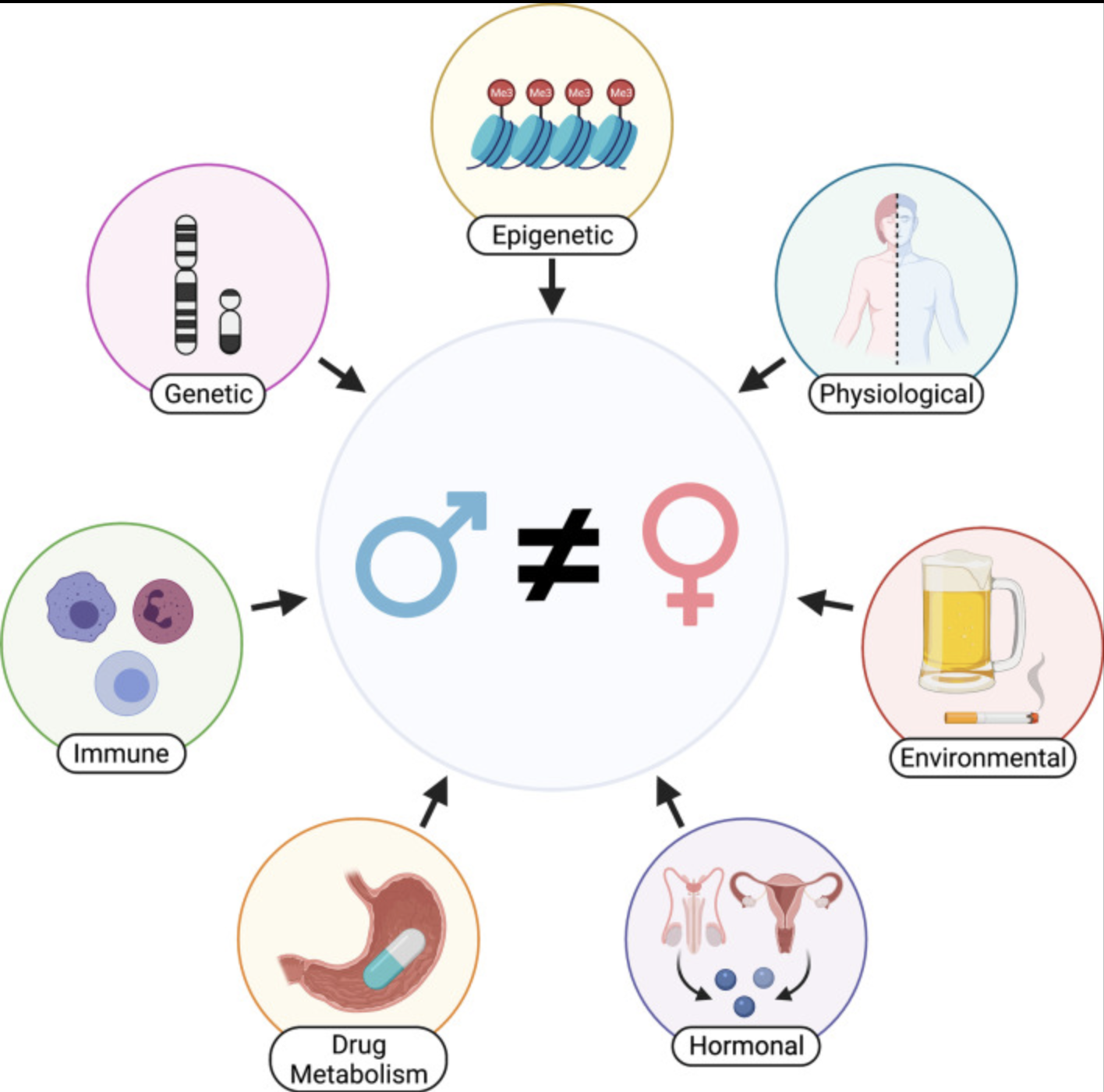
We are dedicated to not only identifying the right drug for the right patient at the right time, but improving that process by prioritizing drug targets and repurposing candidates by assessing sex-biased adverse events, predicting side effects, identifying drugs that will perform similarly in preclinical models and patients, etc.
Recent publications/preprints:
Considerations and challenges for sex-aware drug repurposing, Biology of Sex Differences, 2022.
Methods, Tools, and Approaches for Cross-Condition Studies
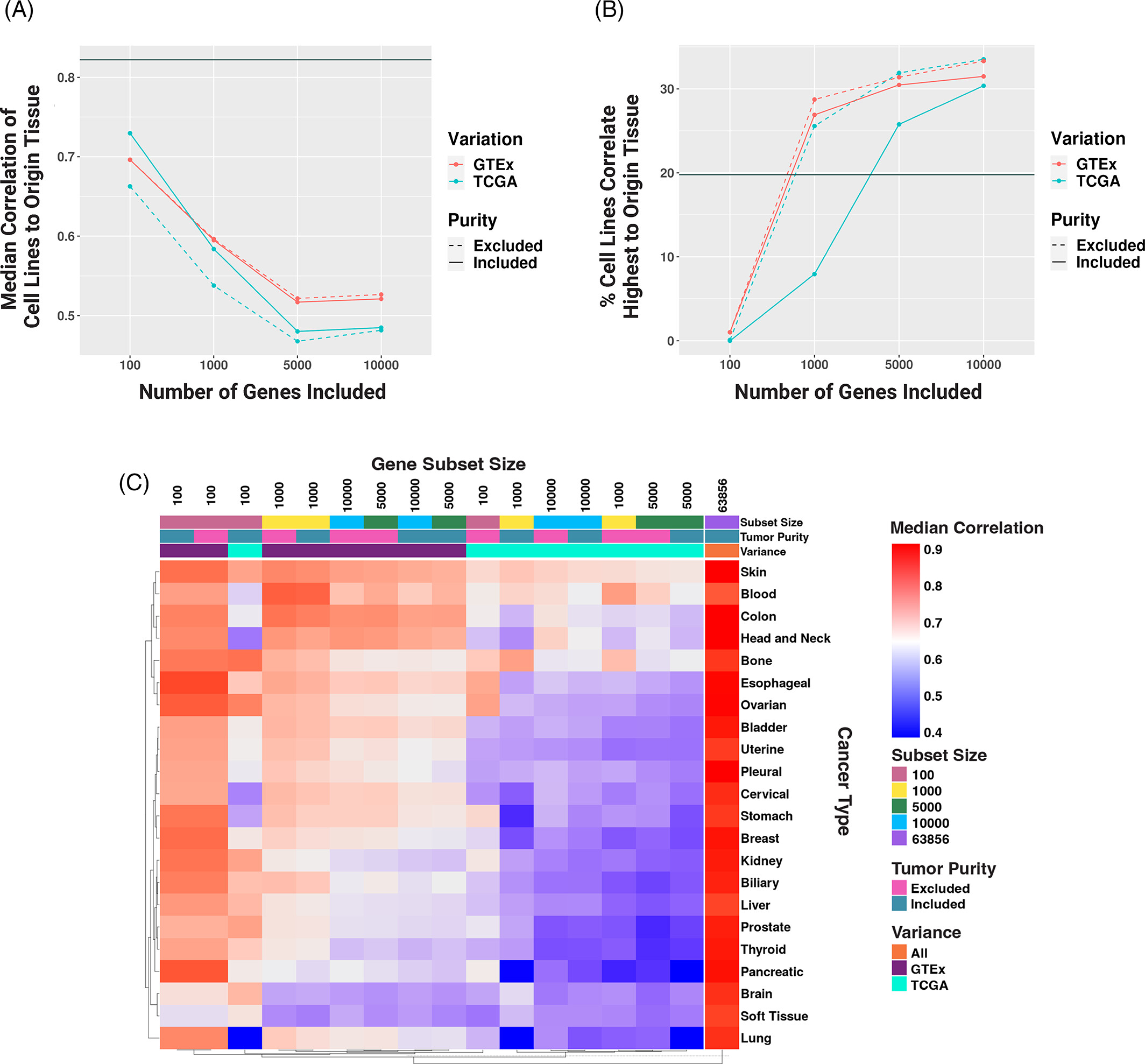
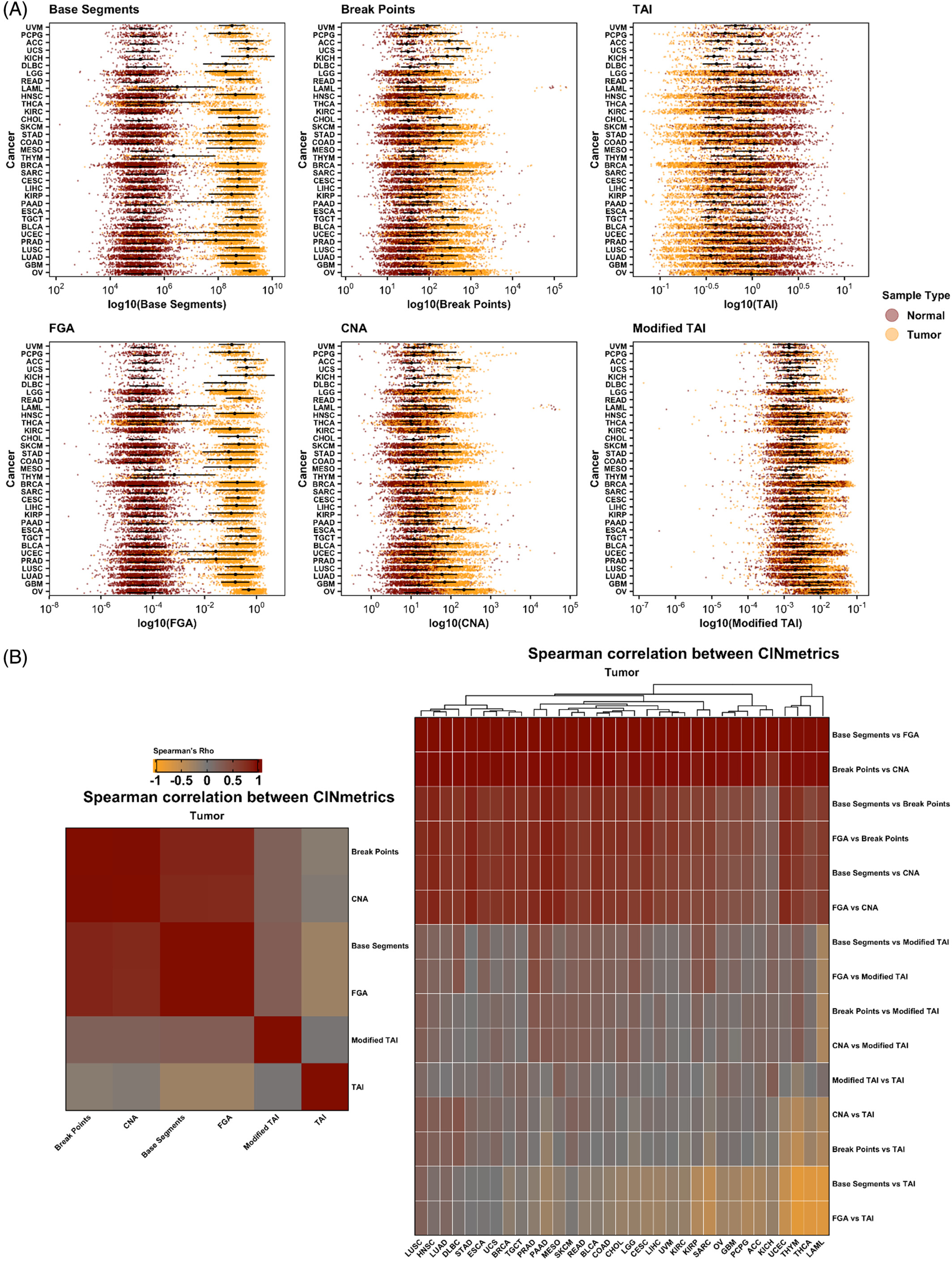
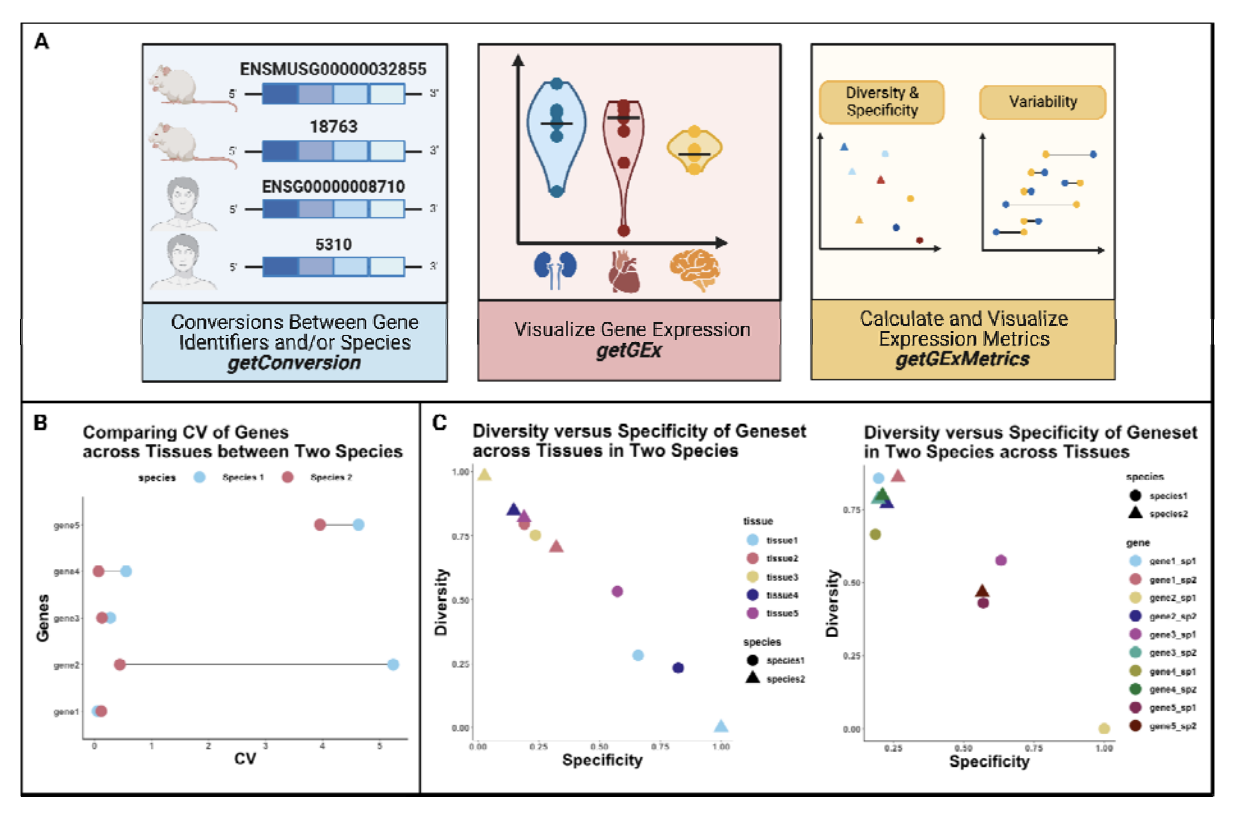
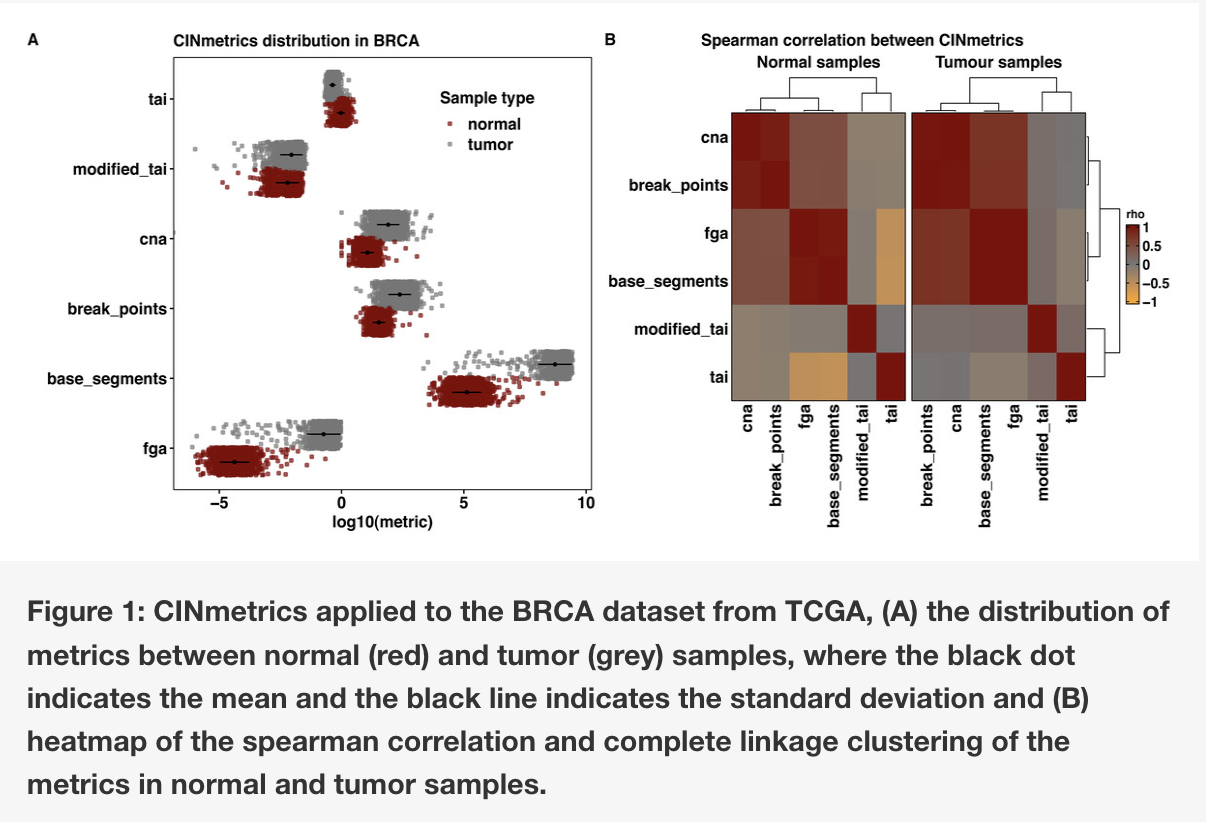
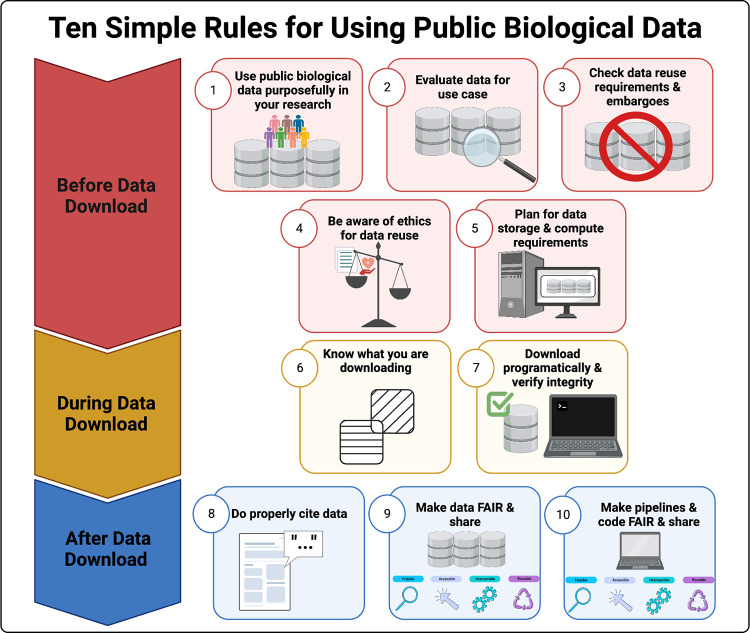
The Lasseigne Lab develops open-source methods, tools, and approaches for studying how extrinsic (e.g., tissue, species) and intrinsic (e.g., transcript expressed, copy number) factors impact disease states and our interpretation of them.
Recent publications/preprints:
Shiny App for the visualization of TF activity in human GTEx data
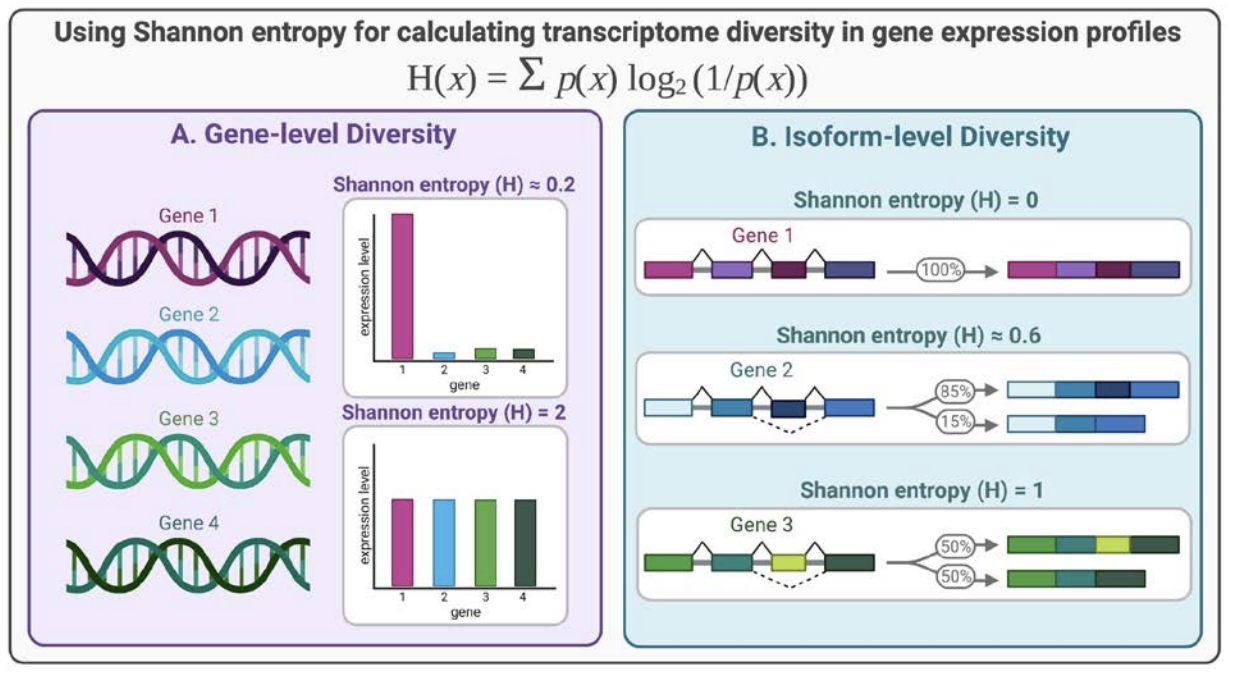
Quantifying transcriptome diversity: a review, Briefings in Functional Genomics, 2023.